Spatially validate scRNA-seq results
Esper High Fidelity Assay – sensitive and specific
Single-cell RNA sequencing (scRNA-seq) has fundamentally expanded our understanding of tissue composition and heterogeneity. While this methodology enables unbiased and comprehensive identification of distinct cell types based on their transcriptomic profiles, it does not provide positional context in the tissue architecture which is critical for understanding the interactions between cells and their native tissue microenvironment.
The Rebus Esper automated platform enables single-molecule RNA Fluorescence In Situ Hybridization (smRNA FISH) chemistry, to map 30+ gene targets across large tissue sections at nanometer resolution. The result is a spatially annotated atlas of embryonic cell types and cell states in the developing human brain.
To dissect cell-type composition and visualize cell clusters in space, a panel of genes identified by dissociated single-cell transcriptomics according to their specificity and expression was interrogated. The full experiment from flow cell assembly to the probing of 30 genes was completed within two days.
The whole human fetal brain, at single mRNA molecules levels of expression, was consistently detected with high signal strength, sensitivity, and specificity using built-in computer vision algorithms. Gene targets were assigned to single nuclei to create a spatially annotated cell x feature matrix for downstream clustering and mapping of cell clusters to XY space.
The results obtained with this robust, automated platform enable validation of functionally distinct cell populations identified by scRNAseq and further reveal the spatial context of developmental gene expression programs at the single cell level. Taken together, these data enable direct observation of area-specific molecular diversity and the ability to infer functional roles of regionally diverse clusters based on known differentiation trajectories and migratory gradients.
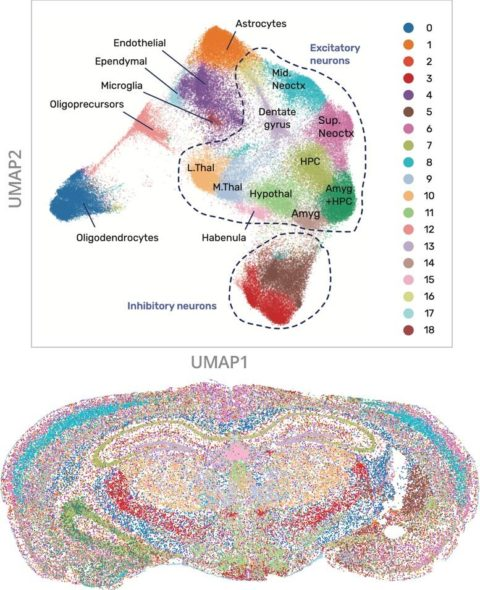
A set of 24 genes – 7 reference and just 17 cell type specific were probed in a coronal section of mouse brain
Our Rebus Esper automated platform rapidly probes 30+ gene targets across large tissue sections at nanometer resolution.


