Technology
Our Technology
Advances in high resolution light microscopy methods are opening exciting new frontiers for direct visualization of biologically important target molecules in their native environment. Quantification of individual molecules of DNA, RNA, and proteins is now possible at the single cell level while preserving important contextual information such as localization and morphology.
The speed limitation of conventional high resolution optical imaging is imposed by the physics of the lens; that is, resolution cannot be improved without dramatically sacrificing field of view (FOV), depth of field (DOF), and working distance (WD) of imaging.
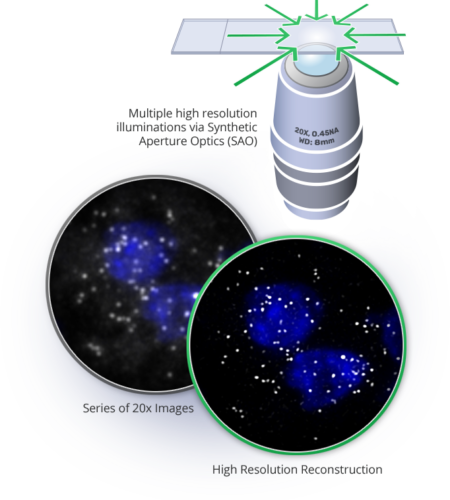
Synthetic Aperture Optics (SAO)
Synthetic Aperture Optics (SAO) is a next-generation fluorescence microscopy solution that improves the lateral resolution of low magnification objectives. The term SAO derives from the fact that illumination from outside of the lens can be used to produce a synthetic Numerical aperture (NA) that is significantly larger than the physical NA of the imaging lens.
SAO decouples resolution from the physics of the lens, allowing higher resolution without sacrificing throughput and ease of use. In SAO, a sample is illuminated by a sequence of high-resolution light patterns formed by the interference of laser beams and a low-resolution lens acquires a series of low resolution images.
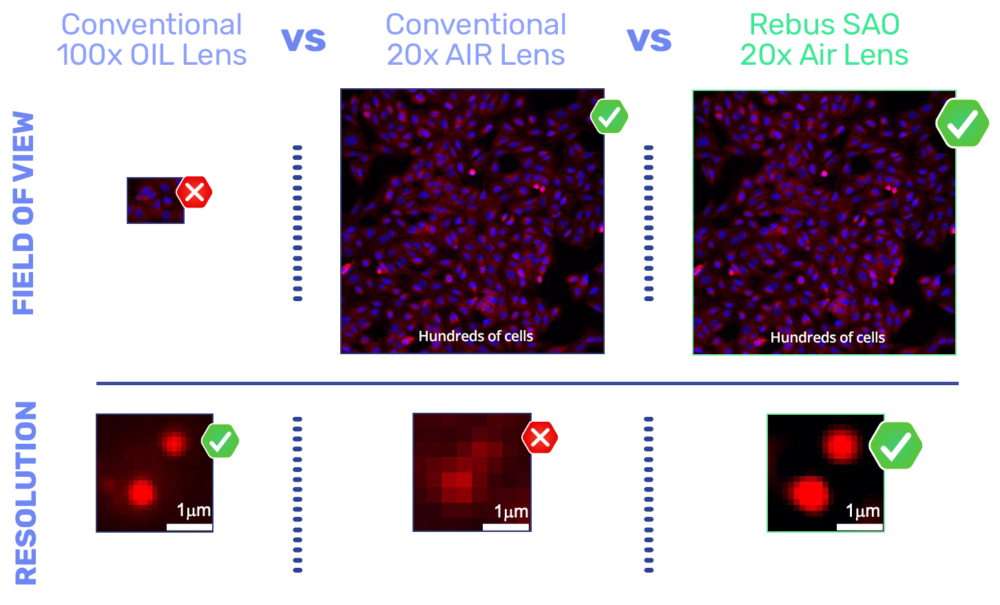
The Rebus Esper spatial omics platform uses SAO to improve a 20x NA 0.45 air lens with similar lateral resolution to a conventional high Numerical aperture (NA) oil immersion lens, enabling more than an order of magnitude improvement in field of view, depth of field, and working distance.
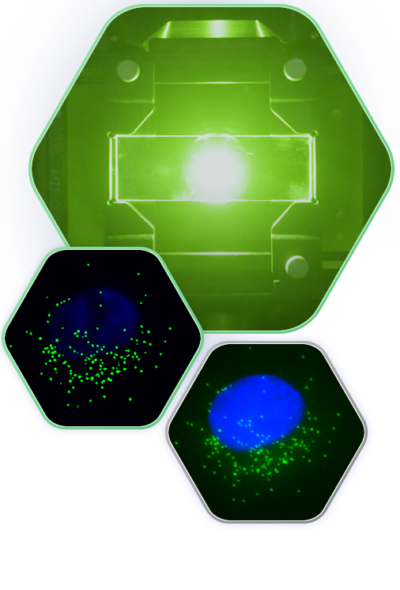
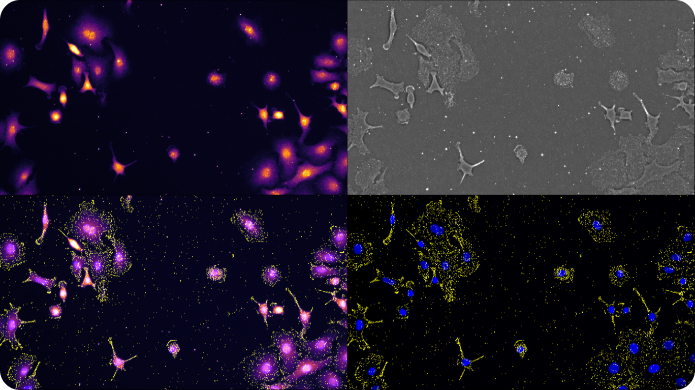
Spot Detection on the Esper™ System
SAO is a powerful imaging modality for extraction of diffraction-limited spots, such as smFISH punctae. During the multi-frame acquisition of each field of view, and as the sample is illuminated from different angles, each spot is turned on in some frames and turned off in others. This property, called modulation, creates a twinkling effect from true smFISH spots when visualizing the animation of frames captured during SAO illumination, and enables Esper to filter true smFISH spots from background noise more efficiently. The pixel size of the reconstructed data is 81nm, meaning that even optically crowded spots can be resolved using a combination of pixel intensity and modulation
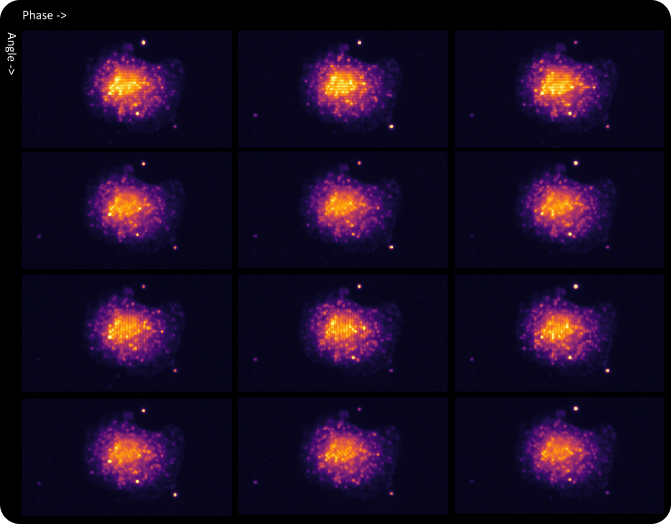
Appearance of a cell with RNA fluorophores in a set of raw images reconstructed by different phase and angle light patterns.
Definition of an smFISH Spot
Single molecule fluorescence in situ hybridization is a strategy for labeling single copies of mRNA for detection with a microscope. Multiple target specific probes hybridize and concentrate fluorescently tagged readout probes to form a spot that is smaller than the wavelength of light or “diffraction limited.”
As the SAO patterns scan through the field of view, these diffraction-limited spots are excited and emit fluorescence when the laser waves intersect with the collection of fluorophores concentrated on the mRNA molecule.
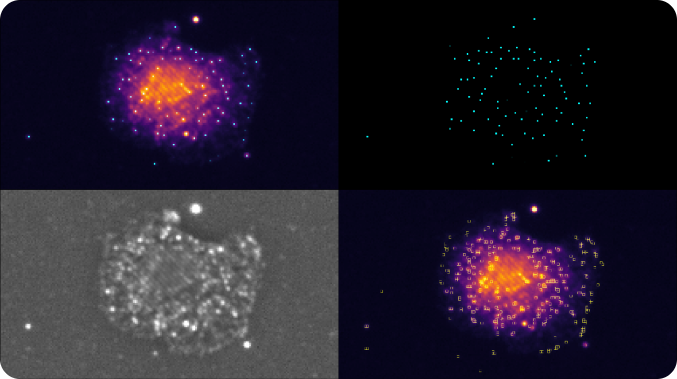
Automatic Spot Extraction Algorithm
Spots generated from single RNA molecules are extracted using a computer vision process that is unique to SAO. The algorithm detects individual fluorophore peaks in the series of raw images for each illumination pattern. Next, the results are combined into a single map per FOV and channel. The peak is measured as a ratio between the peak and the surrounding local background signal level. The higher this metric, the brighter an object appears in the tissue with respect to the surrounding.
The algorithm relies on a small number of parameters to search for the peaks and quantify the background signal. These parameters are modifiable based on the tissue type and can be automatically estimated from the experiment and populated during the processing by the Esper Process software.
DOWNLOAD THE REBUS ESPER BROCHURE